Research
Regulation of chloroplast gene expression.
Chloroplasts are the solar power stations of plant cells. These organelles harbor a network of internal membranes called thylakoids, where the photosynthetic complexes convert light energy into chemical energy. This is then used to synthesize metabolites such as sugars, which are produced from atmospheric carbon dioxide (CO2) and water.
During evolution, plastids have retained a small independent genome derived from their endosymbiotic ancestor, a cyanobacterium. The assembly of the photosynthetic machinery thus requires the coordinate activity of two genomes, in the nucleus and in the chloroplast.

Genetic studies in the green unicellular alga, Chlamydomonas reinhardtii, have revealed many nucleus-encoded proteins which are imported in the chloroplast where they are required for plastid gene expression. They are involved in post-transcriptional steps such RNA splicing, RNA processing or translation (Figure 1). Each of these proteins is surprisingly specific for a single gene in the chloroplast or a small set of genes.
For example, the mRNA of psaA is assembled from three separate transcripts encoded at different loci in the chloroplast genome. The maturation of psaA mRNA requires two steps of splicing in trans which depend on at least fourteen nuclear genes. We have identified some of these genes (Raa1, Raa2, Raa3, Raa7) and shown that the corresponding splicing factors belong to large ribonucleoprotein complexes. In mutants deficient for psaA trans-splicing, the precursor of psaA-exon1 is overexpressed. This overaccumulation is also apparent with chimeric reporter genes under the control of the promoter and 5’untranslated region of psaA-exon1. This over-expression is probably due to the suppression of a negative feed-back loop exerted by the PsaA protein on its own expression (see Wostrikoff et al. (2004) EMBO J).
We are also studying other proteins that are imported into the chloroplast and are necessary for the maturation of psaB mRNA (Mab1), for its translation (Tab2, Tab3), for the maturation of psaC (Mac1) and for the translation of psaA (Taa1) (Figure 1). In response to a change in the growth medium, such as iron starvation, we observed a concerted decrease in the amount of the subunits of PSI and of the Taa1 and Mac1 proteins. Furthermore the degree of phosphorylation of Mac1 also responds to changes in environmental conditions, suggesting a regulatory mechanism.
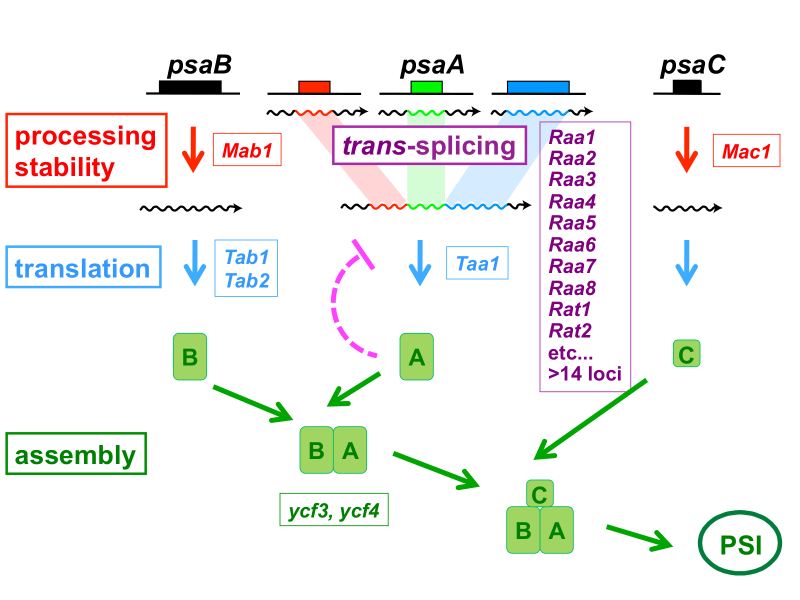
Figure 1: Genetic dissection of photosystem I biogenesis
Light acclimation of the photosynthetic machinery.
In the photosynthetic electron transfer chain of the chloroplast thylakoid membranes, two photosystems (PSII and PSI) work in series to produce reducing power (NADPH) and energy (ATP), which are used for metabolic processes such as CO2 fixation. Depending on the quality and intensity of light, part of the light harvesting antenna LHCII can associate with either PSII or PSI (Figure 2). This balances the activity of the two photosystems and ensures optimal activity of the photosynthetic chain. This regulatory mechanism, called state transition, also responds to the metabolic requirements of the cell by modulating cyclic electron flow around PSI, which allows the production of ATP without NADPH. State transitions and the architecture of thylakoid membranes are regulated by protein kinases (Stt7 and Stl1 in Chlamydomonas, STN7 and STN8 in Arabidopsis) that phosphorylate key components of LHCII and of PSII. We have recently identified specific phosphatases, PPH1/TAP38 and PBCP, that dephosphorylate these proteins and thus contribute to the reversibility of these regulatory mechanisms.
Upon exposure to excess light, algae and plants can dissipate part of the captured light energy as heat. In collaboration with the group of Prof. Roman Ulm in our Department, we have shown that perception of UV-B by the photoreceptor UVR8 of Chlamydomonas also induces the expression of genes involved in thermal dissipation of light energy (qE), in particular PSBS and LHCSR1.
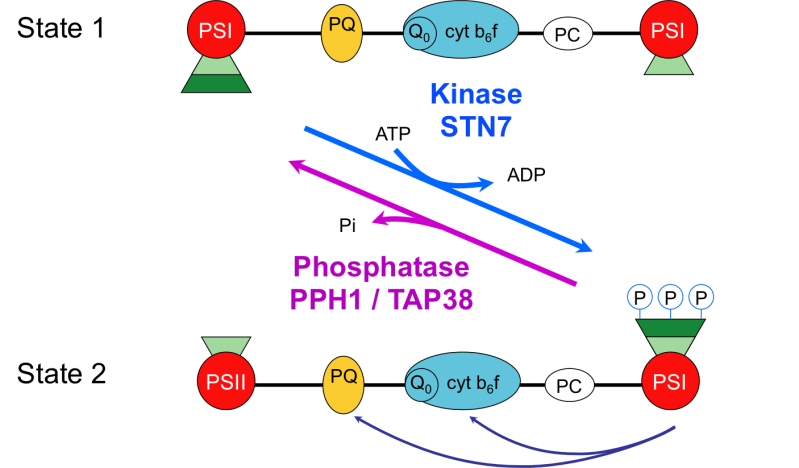
Figure 2: State transitions
